── Attaching packages ───────────────────────────── tidyverse 1.3.1 ──✓ ggplot2 3.3.5 ✓ purrr 0.3.4
✓ tibble 3.1.4 ✓ dplyr 1.0.7
✓ tidyr 1.1.3 ✓ stringr 1.4.0
✓ readr 2.0.1 ✓ forcats 0.5.1── Conflicts ──────────────────────────────── tidyverse_conflicts() ──
x dplyr::filter() masks stats::filter()
x dplyr::lag() masks stats::lag()Loading required package: RcppLoading 'brms' package (version 2.16.2). Useful instructions
can be found by typing help('brms'). A more detailed introduction
to the package is available through vignette('brms_overview').
Attaching package: 'brms'The following object is masked from 'package:stats':
arplot_curve <- function(model, method = "fitted") {
cond <- tibble(.rows = 1)
cond[[names(model$data)[2]]] <- "none"
plot(conditional_effects(model, method = method,
conditions = cond,
spaghetti = TRUE, nsamples = 100),
points = TRUE,
point_args = list(alpha = 0.05),
spaghetti_args = list(alpha = 0.01, colour = '#FF000007'),
line_args = list(size = 0),
plot = FALSE)[[1]]
}
rmse <- function(error) { sqrt(mean(error^2, na.rm = TRUE)) }
rmse_brms <- function(model) {
resid <- residuals(model, summary = F)
rmse_samples <- apply(resid, 1, rmse)
rmse_ci <- rstantools::posterior_interval(as.matrix(rmse_samples), prob = 0.95)
sprintf("%.2f [%.2f;%.2f]", mean(rmse_samples), rmse_ci[1], rmse_ci[2])
}
mae <- function(error) { mean(abs(error), na.rm = TRUE) }
mae_brms <- function(model) {
resid <- residuals(model, summary = F)
mae_samples <- apply(resid, 1, mae)
mae_ci <- rstantools::posterior_interval(as.matrix(mae_samples), prob = 0.95)
sprintf("%.2f [%.2f;%.2f]", mean(mae_samples), mae_ci[1], mae_ci[2])
}
loo_ci <- function(model) {
loo_R <- loo_R2(model, re_formula = NA)
loo_R <- sqrt(loo_R)
sprintf("%.2f [%.2f;%.2f]", loo_R[,"Estimate"], loo_R[,"Q2.5"], loo_R[,"Q97.5"])
}
biocycle <- readRDS("biocycle.rds")
options(mc.cores = parallel::detectCores(), brms.backend = "cmdstanr", brms.file_refit = "on_change")
rstan::rstan_options(auto_write = TRUE)
biocycle <- biocycle %>% filter(between(cycle_length, 20, 35))
biocycle$logE_P <- log(biocycle$E_P)
biocycle$logE <- log(biocycle$total_estradiol)
biocycle$logP <- log(biocycle$progesterone)
biocycle$logT <- log(biocycle$Testo)
biocycle$logFE <- log(biocycle$estradiol)
biocycle$logFT <- log(biocycle$`Free T (ng/dL)`)
biocycle$fertile_lh <- NULL
Days
bc_days <- data.frame(
bc_day = c(-28:-1, -29:-40),
prc_stirn_bc = c(.01, .01, .02, .03, .05, .09, .16, .27, .38, .48, .56, .58, .55, .48, .38, .28, .20, .14, .10, .07, .06, .04, .03, .02, .01, .01, .01, .01, rep(.01, times = 12)),
# rep(.01, times = 70)), # gangestad uses .01 here, but I think such cases are better thrown than kept, since we might simply have missed a mens
prc_wcx_bc = c(.000, .000, .001, .002, .004, .009, .018, .032, .050, .069, .085, .094, .093, .085, .073, .059, .047, .036, .028, .021, .016, .013, .010, .008, .007, .006, .005, .005, rep(.005, times = 12))
) %>% arrange(bc_day)
fc_days <- data.frame(
fc_day = c(0:39),
prc_stirn_fc = c(.01, .01, .02, .03, .05, .09, .16, .27, .38, .48, .56, .58, .55, .48, .38, .28, .20, .14, .10, .07, .06, .04, .03, .02, .01, .01, .01, .01, rep(.01, times = 12)),
# rep(.01, times = 70)), # gangestad uses .01 here, but I think such cases are better thrown than kept, since we might simply have missed a mens
prc_wcx_fc = c(.000, .000, .001, .002, .004, .009, .018, .032, .050, .069, .085, .094, .093, .085, .073, .059, .047, .036, .028, .021, .016, .013, .010, .008, .007, .006, .005, .005, rep(.005, times = 12))
)
lh_days <- tibble(
conception_risk_lh = c(rep(0,8), 0.00, 0.01, 0.02, 0.06, 0.16, 0.20, 0.25, 0.24, 0.10, 0.02, 0.02, rep(0,12)),
lh_day = -15:15
) %>%
mutate(fertile_lh = conception_risk_lh/max(conception_risk_lh)*0.25/0.31)
biocycle <- biocycle %>% left_join(lh_days %>% select(lh_day, fertile_lh), by = "lh_day")
Impute E
Backward counting
(mod_e_bc = brm(bf(logE | cens(estradiol_cens) ~ s(bc_day) + (1 | id) + (1|id:cycle)),
data = biocycle, control = list(adapt_delta = 0.95, max_treedepth = 20),
file ='models/mod_e_bc'))
Warning: Rows containing NAs were excluded from the model. Family: gaussian
Links: mu = identity; sigma = identity
Formula: logE | cens(estradiol_cens) ~ s(bc_day) + (1 | id) + (1 | id:cycle)
Data: biocycle (Number of observations: 3425)
Draws: 4 chains, each with iter = 1000; warmup = 0; thin = 1;
total post-warmup draws = 4000
Smooth Terms:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sds(sbc_day_1) 5.18 1.26 3.29 8.24 1.00 791
Tail_ESS
sds(sbc_day_1) 1306
Group-Level Effects:
~id (Number of levels: 246)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sd(Intercept) 0.29 0.02 0.25 0.32 1.00 1304
Tail_ESS
sd(Intercept) 2659
~id:cycle (Number of levels: 448)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sd(Intercept) 0.07 0.03 0.01 0.12 1.01 317
Tail_ESS
sd(Intercept) 601
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 4.45 0.02 4.41 4.49 1.00 1054 1721
sbc_day_1 -13.26 1.71 -16.66 -9.89 1.00 2509 2890
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 0.49 0.01 0.48 0.51 1.00 3859 3058
Draws were sampled using sample(hmc). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).performance::variance_decomposition(mod_e_bc)
# Random Effect Variances and ICC
Conditioned on: all random effects
## Variance Ratio (comparable to ICC)
Ratio: 0.14 CI 95%: [0.09 0.19]
## Variances of Posterior Predicted Distribution
Conditioned on fixed effects: 0.53 CI 95%: [0.50 0.56]
Conditioned on rand. effects: 0.61 CI 95%: [0.58 0.64]
## Difference in Variances
Difference: 0.08 CI 95%: [0.05 0.12]# performance::model_performance(mod_e_bc, metrics = c("RMSE", "R2", "R2_adj"))
loo_R2(mod_e_bc, re_formula = NA)
Warning: Results may not be meaningful for censored models. Estimate Est.Error Q2.5 Q97.5
R2 0.4590614 0.01175674 0.4353065 0.481754rmse_brms(mod_e_bc)
Warning: Results may not be meaningful for censored models.[1] "0.49 [0.48;0.49]"mae_brms(mod_e_bc)
Warning: Results may not be meaningful for censored models.[1] "0.39 [0.38;0.39]"plot_curve(mod_e_bc)
Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.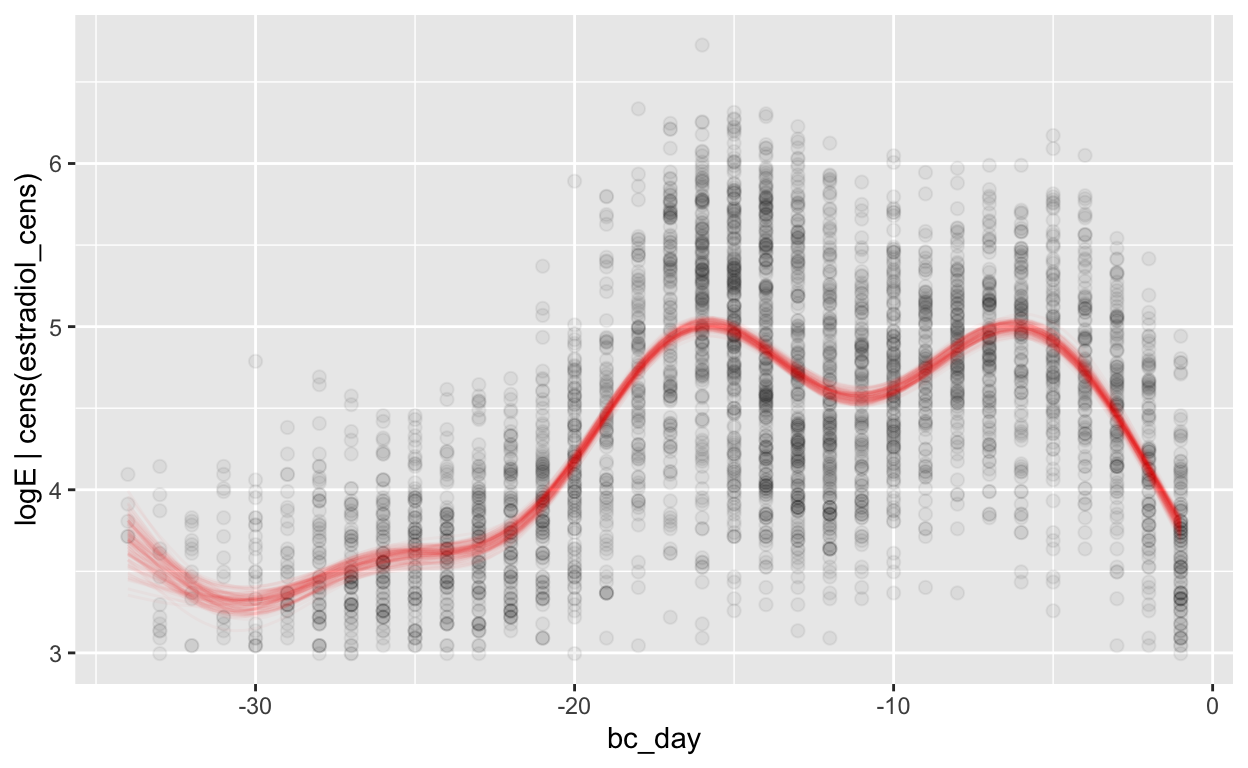
Forward counting
(mod_e_fc = brm(bf(logE | cens(estradiol_cens) ~ s(fc_day) + (1 | id) + (1|id:cycle)),
data = biocycle, control = list(adapt_delta = 0.95, max_treedepth = 20),
file ='models/mod_e_fc'))
Warning: Rows containing NAs were excluded from the model. Family: gaussian
Links: mu = identity; sigma = identity
Formula: logE | cens(estradiol_cens) ~ s(fc_day) + (1 | id) + (1 | id:cycle)
Data: biocycle (Number of observations: 3425)
Draws: 4 chains, each with iter = 1000; warmup = 0; thin = 1;
total post-warmup draws = 4000
Smooth Terms:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sds(sfc_day_1) 2.00 0.61 1.12 3.43 1.00 1260
Tail_ESS
sds(sfc_day_1) 2183
Group-Level Effects:
~id (Number of levels: 246)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sd(Intercept) 0.29 0.02 0.26 0.33 1.00 997
Tail_ESS
sd(Intercept) 2069
~id:cycle (Number of levels: 448)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sd(Intercept) 0.03 0.02 0.00 0.08 1.01 398
Tail_ESS
sd(Intercept) 983
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 4.45 0.02 4.41 4.49 1.00 1192 2141
sfc_day_1 -1.03 1.61 -4.22 2.16 1.00 1753 2502
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 0.57 0.01 0.55 0.58 1.00 4618 2866
Draws were sampled using sample(hmc). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).loo_R2(mod_e_fc, re_formula = NA)
Warning: Results may not be meaningful for censored models. Estimate Est.Error Q2.5 Q97.5
R2 0.3293852 0.01217784 0.3053314 0.3523772rmse_brms(mod_e_fc)
Warning: Results may not be meaningful for censored models.[1] "0.56 [0.56;0.56]"mae_brms(mod_e_fc)
Warning: Results may not be meaningful for censored models.[1] "0.45 [0.45;0.46]"plot_curve(mod_e_fc, method = "fitted")
Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.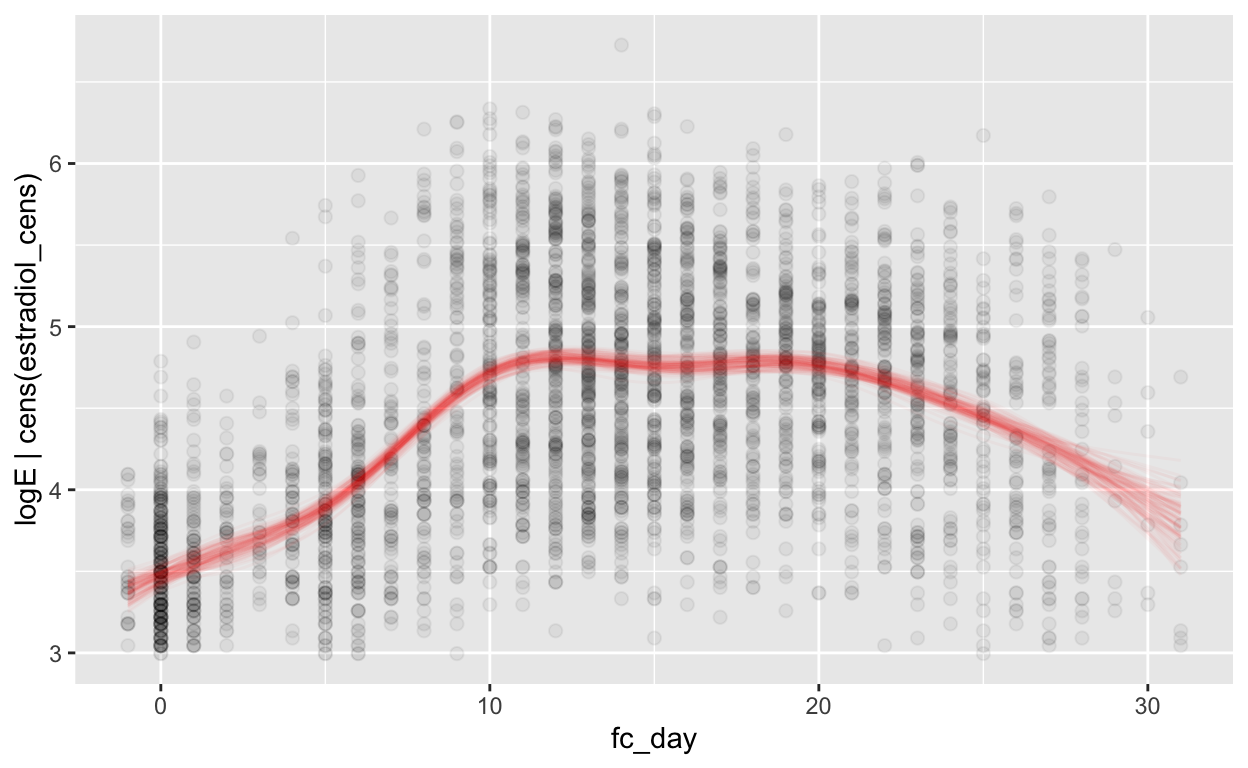
Relative to LH surge
(mod_e_lh = brm(bf(logE | cens(estradiol_cens) ~ s(lh_day) + (1 | id) + (1|id:cycle)),
data = biocycle %>% filter(between(lh_day, -20, 14)), control = list(adapt_delta = 0.95, max_treedepth = 20),
file ='models/mod_e_lh')
)
Warning: Rows containing NAs were excluded from the model. Family: gaussian
Links: mu = identity; sigma = identity
Formula: logE | cens(estradiol_cens) ~ s(lh_day) + (1 | id) + (1 | id:cycle)
Data: biocycle %>% filter(between(lh_day, -20, 14)) (Number of observations: 2590)
Draws: 4 chains, each with iter = 1000; warmup = 0; thin = 1;
total post-warmup draws = 4000
Smooth Terms:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sds(slh_day_1) 8.38 2.01 5.54 13.26 1.00 1026
Tail_ESS
sds(slh_day_1) 1645
Group-Level Effects:
~id (Number of levels: 214)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sd(Intercept) 0.27 0.02 0.24 0.31 1.00 1009
Tail_ESS
sd(Intercept) 2117
~id:cycle (Number of levels: 340)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sd(Intercept) 0.07 0.03 0.01 0.12 1.02 288
Tail_ESS
sd(Intercept) 576
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 4.46 0.02 4.42 4.51 1.00 1180 1664
slh_day_1 0.12 2.06 -3.87 4.27 1.00 2186 2502
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 0.46 0.01 0.45 0.47 1.00 4236 3057
Draws were sampled using sample(hmc). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).loo_R2(mod_e_lh, re_formula = NA)
Warning: Results may not be meaningful for censored models. Estimate Est.Error Q2.5 Q97.5
R2 0.5182738 0.01413739 0.4900016 0.5460118rmse_brms(mod_e_lh)
Warning: Results may not be meaningful for censored models.[1] "0.45 [0.45;0.46]"mae_brms(mod_e_lh)
Warning: Results may not be meaningful for censored models.[1] "0.36 [0.35;0.36]"plot_curve(mod_e_lh, method = "fitted")
Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.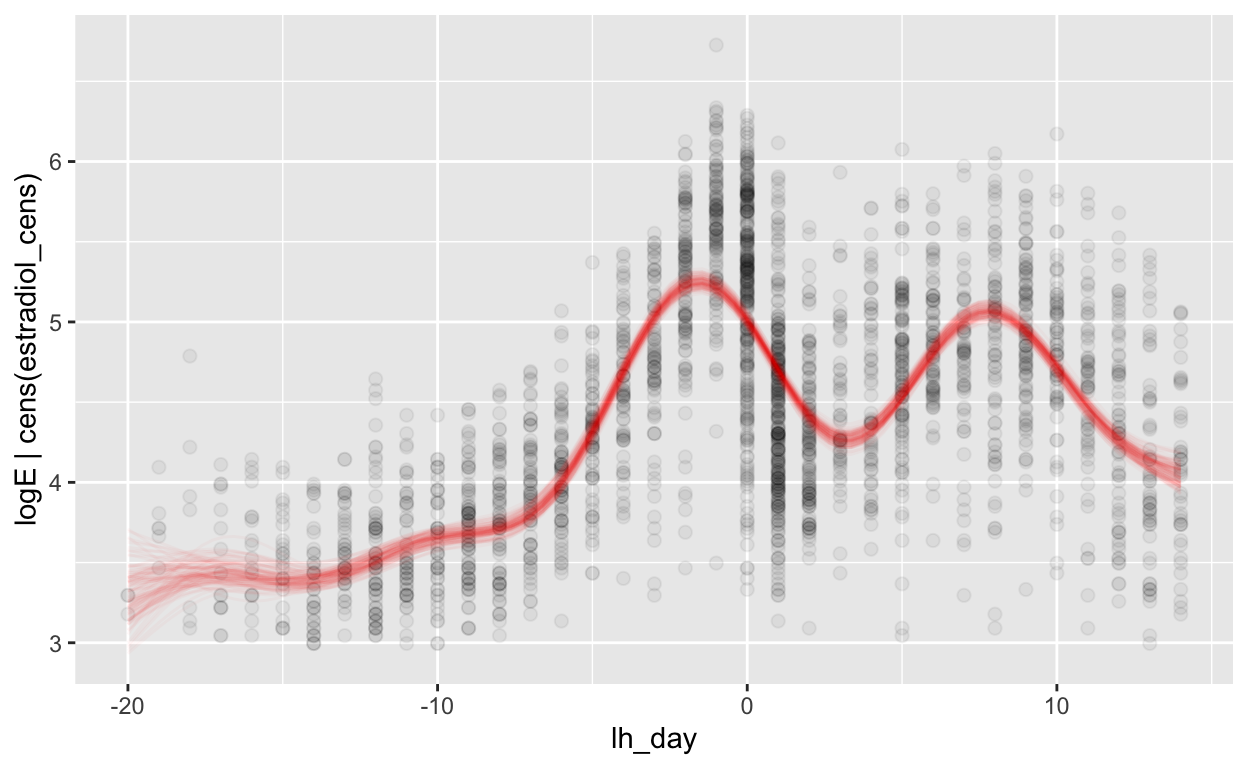
Impute FE
Backward counting
(mod_fe_bc = brm(bf(logFE | cens(estradiol_cens) ~ s(bc_day) + (1 | id) + (1|id:cycle)),
data = biocycle, control = list(adapt_delta = 0.95, max_treedepth = 20),
file ='models/mod_fe_bc'))
Warning: Rows containing NAs were excluded from the model. Family: gaussian
Links: mu = identity; sigma = identity
Formula: logFE | cens(estradiol_cens) ~ s(bc_day) + (1 | id) + (1 | id:cycle)
Data: biocycle (Number of observations: 3425)
Draws: 4 chains, each with iter = 1000; warmup = 0; thin = 1;
total post-warmup draws = 4000
Smooth Terms:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sds(sbc_day_1) 5.89 1.58 3.70 9.72 1.00 952
Tail_ESS
sds(sbc_day_1) 1290
Group-Level Effects:
~id (Number of levels: 246)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sd(Intercept) 0.28 0.02 0.25 0.32 1.00 1096
Tail_ESS
sd(Intercept) 2224
~id:cycle (Number of levels: 448)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sd(Intercept) 0.08 0.03 0.01 0.14 1.01 343
Tail_ESS
sd(Intercept) 368
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Intercept 0.42 0.02 0.38 0.46 1.00 1624 2302
sbc_day_1 -14.50 1.98 -18.44 -10.56 1.00 2834 2460
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma 0.56 0.01 0.54 0.57 1.00 3912 2790
Draws were sampled using sample(hmc). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).loo_R2(mod_fe_bc, re_formula = NA)
Warning: Results may not be meaningful for censored models. Estimate Est.Error Q2.5 Q97.5
R2 0.4575188 0.01199411 0.4334088 0.4797044rmse_brms(mod_fe_bc)
Warning: Results may not be meaningful for censored models.[1] "0.55 [0.54;0.55]"mae_brms(mod_fe_bc)
Warning: Results may not be meaningful for censored models.[1] "0.43 [0.43;0.44]"plot_curve(mod_fe_bc, method = "fitted")
Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.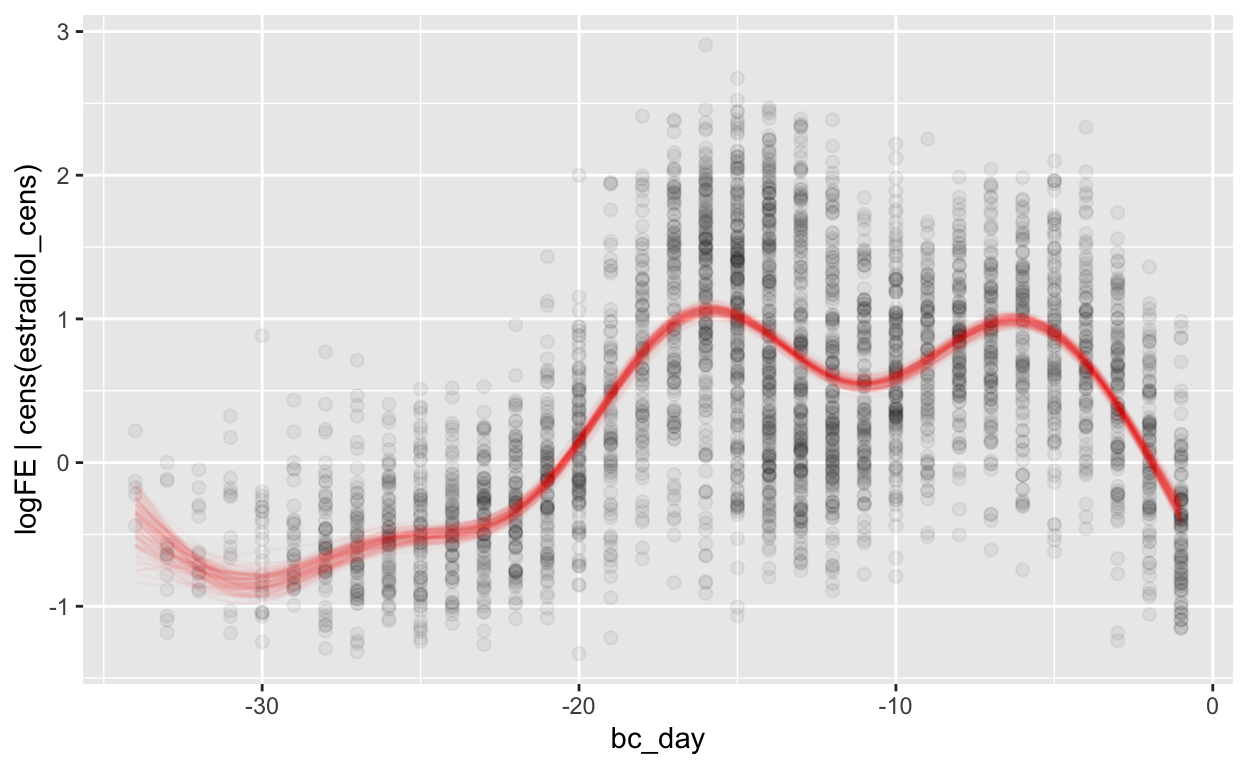
Forward counting
mod_fe_fc = brm(bf(logFE | cens(estradiol_cens) ~ s(fc_day) + (1 | id) + (1|id:cycle)),
data = biocycle, control = list(adapt_delta = 0.95, max_treedepth = 20),
file ='models/mod_fe_fc')
Warning: Rows containing NAs were excluded from the model.loo_R2(mod_fe_fc, re_formula = NA)
Warning: Results may not be meaningful for censored models. Estimate Est.Error Q2.5 Q97.5
R2 0.3296142 0.01229912 0.305068 0.3526868rmse_brms(mod_fe_fc)
Warning: Results may not be meaningful for censored models.[1] "0.63 [0.63;0.63]"mae_brms(mod_fe_fc)
Warning: Results may not be meaningful for censored models.[1] "0.51 [0.50;0.51]"plot_curve(mod_fe_fc, method = "fitted")
Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.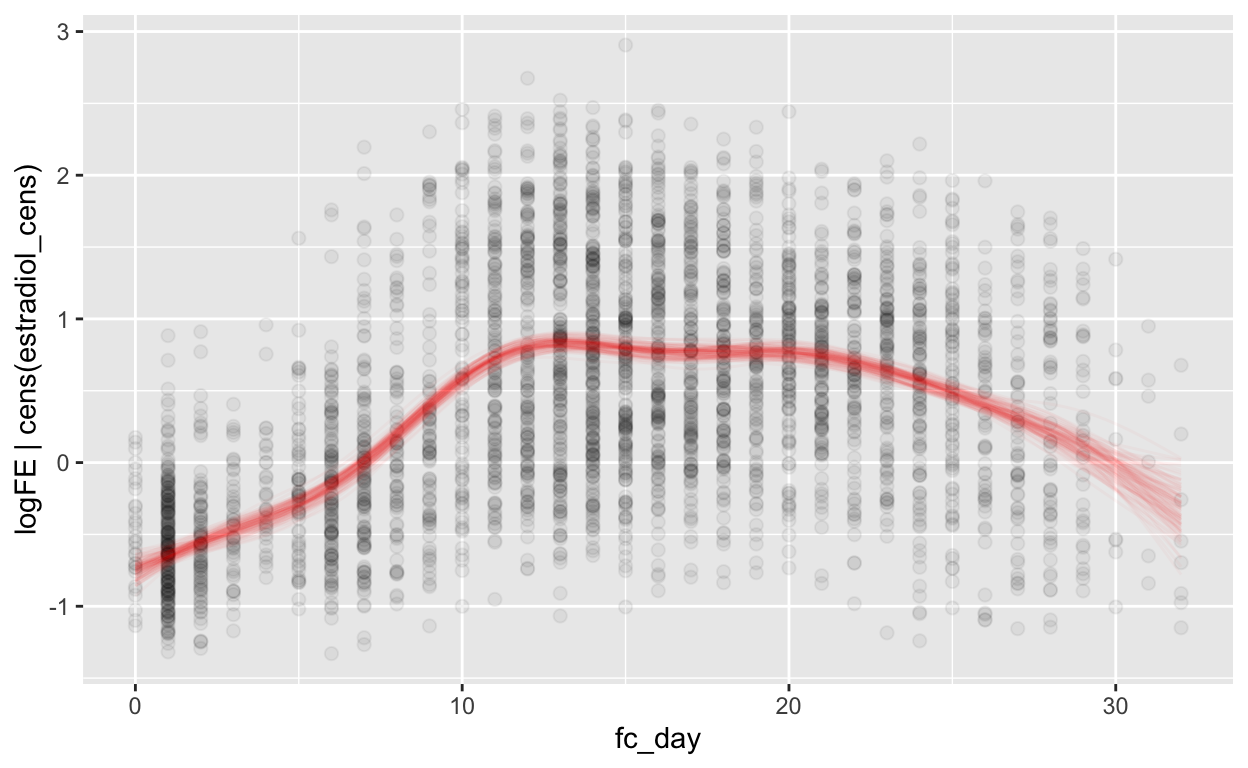
Relative to LH surge
mod_fe_lh = brm(bf(logFE | cens(estradiol_cens) ~ s(lh_day) + (1 | id) + (1|id:cycle)),
data = biocycle %>% filter(between(lh_day, -20, 14)), control = list(adapt_delta = 0.95, max_treedepth = 20),
file ='models/mod_fe_lh')
Warning: Rows containing NAs were excluded from the model.loo_R2(mod_fe_lh, re_formula = NA)
Warning: Results may not be meaningful for censored models. Estimate Est.Error Q2.5 Q97.5
R2 0.5213177 0.01417518 0.4931336 0.5481039rmse_brms(mod_fe_lh)
Warning: Results may not be meaningful for censored models.[1] "0.51 [0.50;0.51]"mae_brms(mod_fe_lh)
Warning: Results may not be meaningful for censored models.[1] "0.40 [0.39;0.40]"plot_curve(mod_fe_lh, method = "fitted")
Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.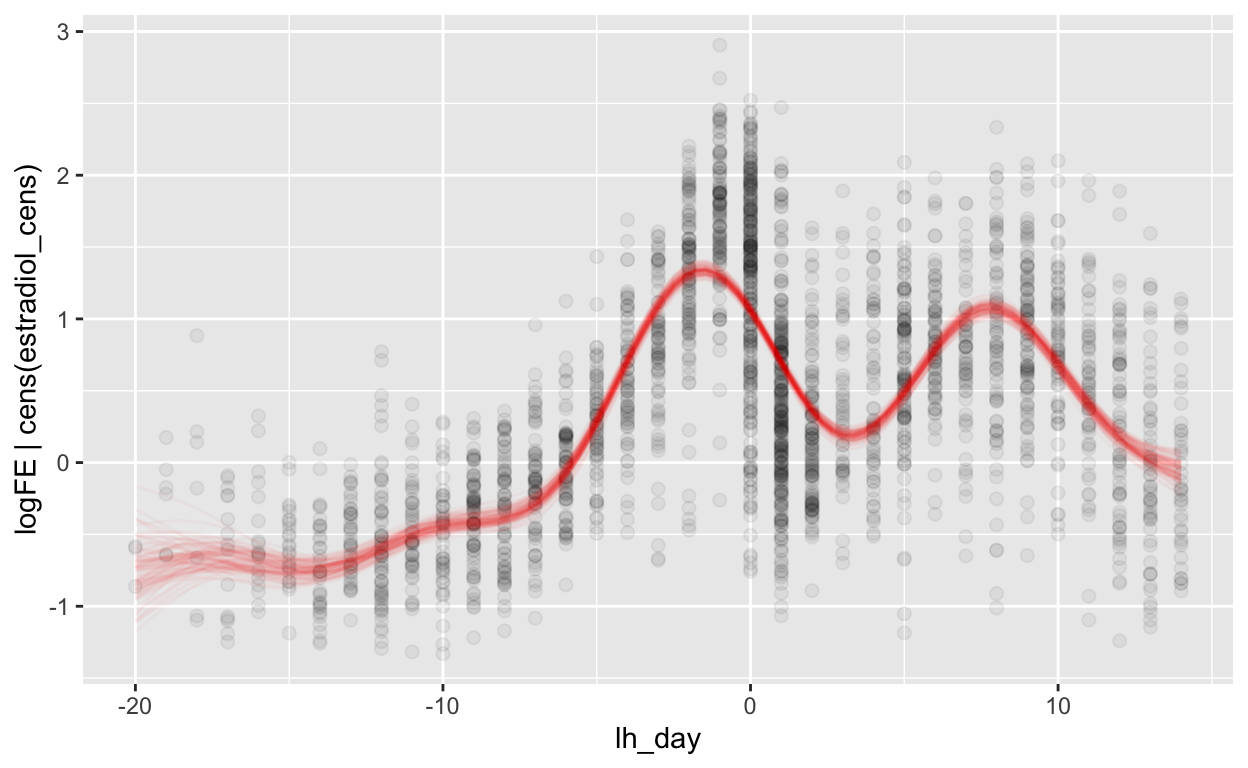
Impute P
Backward counting
mod_p_bc = brm(bf(logP | cens(progesterone_cens) ~ s(bc_day) + (1 | id) + (1|id:cycle)),
data = biocycle, control = list(adapt_delta = 0.95, max_treedepth = 20),
file ='models/mod_p_bc')
Warning: Rows containing NAs were excluded from the model.plot_curve(mod_p_bc, method = "fitted")
Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.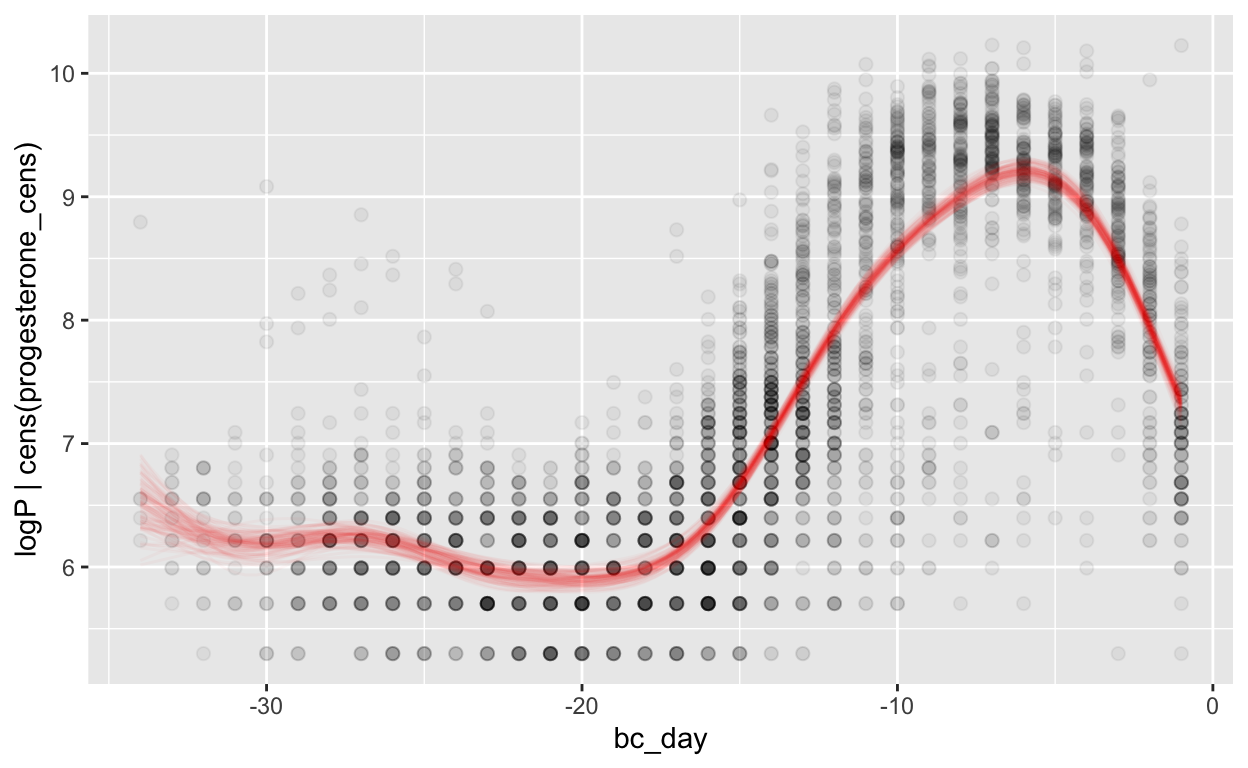
Forward counting
mod_p_fc = brm(bf(logP | cens(progesterone_cens) ~ s(fc_day) + (1 | id) + (1|id:cycle)),
data = biocycle, control = list(adapt_delta = 0.95, max_treedepth = 20),
file ='models/mod_p_fc')
Warning: Rows containing NAs were excluded from the model.plot_curve(mod_p_fc, method = "fitted")
Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.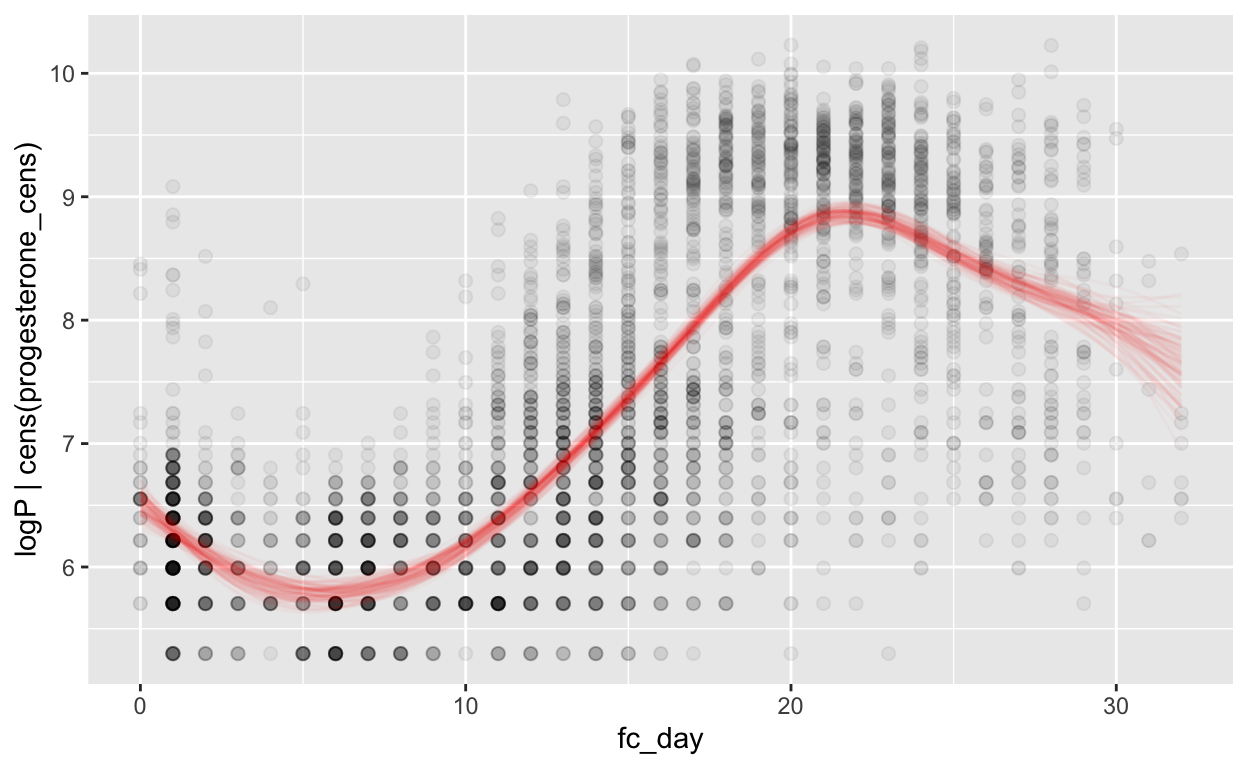
Relative to LH surge
mod_p_lh = brm(bf(logP | cens(progesterone_cens) ~ s(lh_day) + (1 | id) + (1|id:cycle)),
data = biocycle %>% filter(between(lh_day, -20, 14)), control = list(adapt_delta = 0.95, max_treedepth = 20),
file ='models/mod_p_lh')
Warning: Rows containing NAs were excluded from the model.plot_curve(mod_p_lh, method = "fitted")
Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.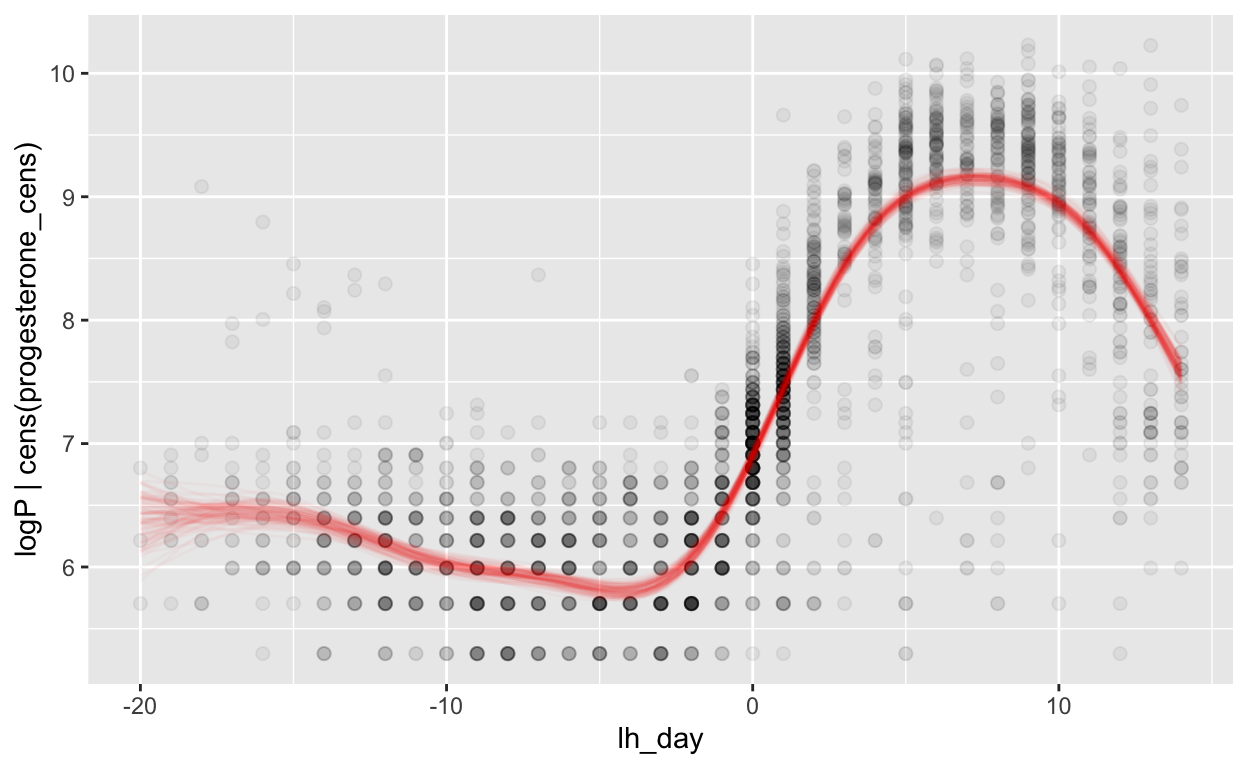
New PBFW (probability of being in fertile window)
Backward counting
mod_pbfw_bc = brm(bf(fertile_lh ~ s(bc_day) + (1 | id) + (1|id:cycle)),
data = biocycle,
file ='models/mod_pbfw_bc')
Warning: Rows containing NAs were excluded from the model.plot_curve(mod_pbfw_bc, method = "fitted")
Warning: The following variables in 'conditions' are not part of the model:
'id'Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.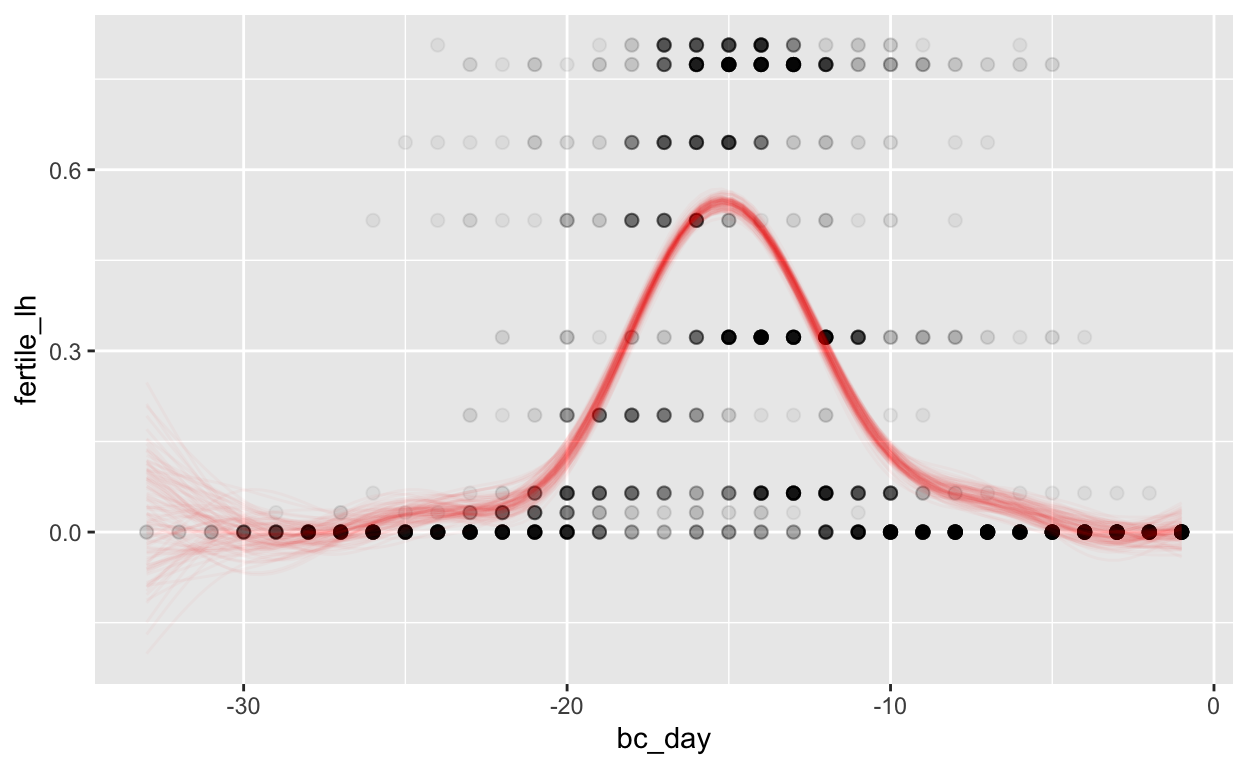
Forward counting
mod_pbfw_fc = brm(bf(fertile_lh ~ s(fc_day) + (1 | id) + (1|id:cycle)),
data = biocycle,
file ='models/mod_pbfw_fc')
Warning: Rows containing NAs were excluded from the model.plot_curve(mod_pbfw_fc, method = "fitted")
Warning: The following variables in 'conditions' are not part of the model:
'id'Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.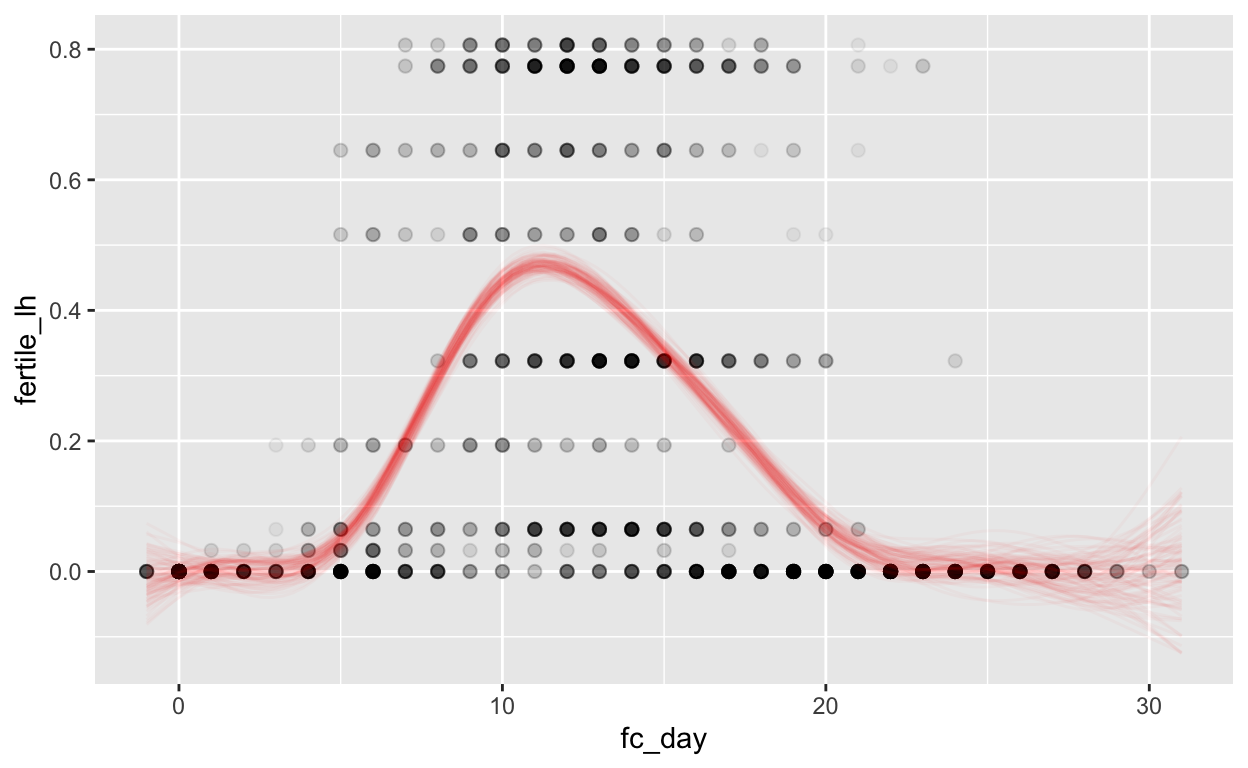
Relative to LH surge
Not done, because predictor = criterion.
Impute T
Backward counting
mod_t_bc = brm(bf(logT | cens(Testo_cens) ~ s(bc_day) + (1 | id) + (1|id:cycle)),
data = biocycle, control = list(adapt_delta = 0.95, max_treedepth = 20),
file ='models/mod_t_bc')
Warning: Rows containing NAs were excluded from the model.loo_R2(mod_t_bc, re_formula = NA)
Warning: Results may not be meaningful for censored models.Warning: Some Pareto k diagnostic values are too high. See help('pareto-k-diagnostic') for details. Estimate Est.Error Q2.5 Q97.5
R2 0.05955607 0.01125779 0.03759546 0.08188889rmse_brms(mod_t_bc)
Warning: Results may not be meaningful for censored models.[1] "0.17 [0.17;0.18]"mae_brms(mod_t_bc)
Warning: Results may not be meaningful for censored models.[1] "0.13 [0.13;0.13]"plot_curve(mod_t_bc, method = "fitted")
Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.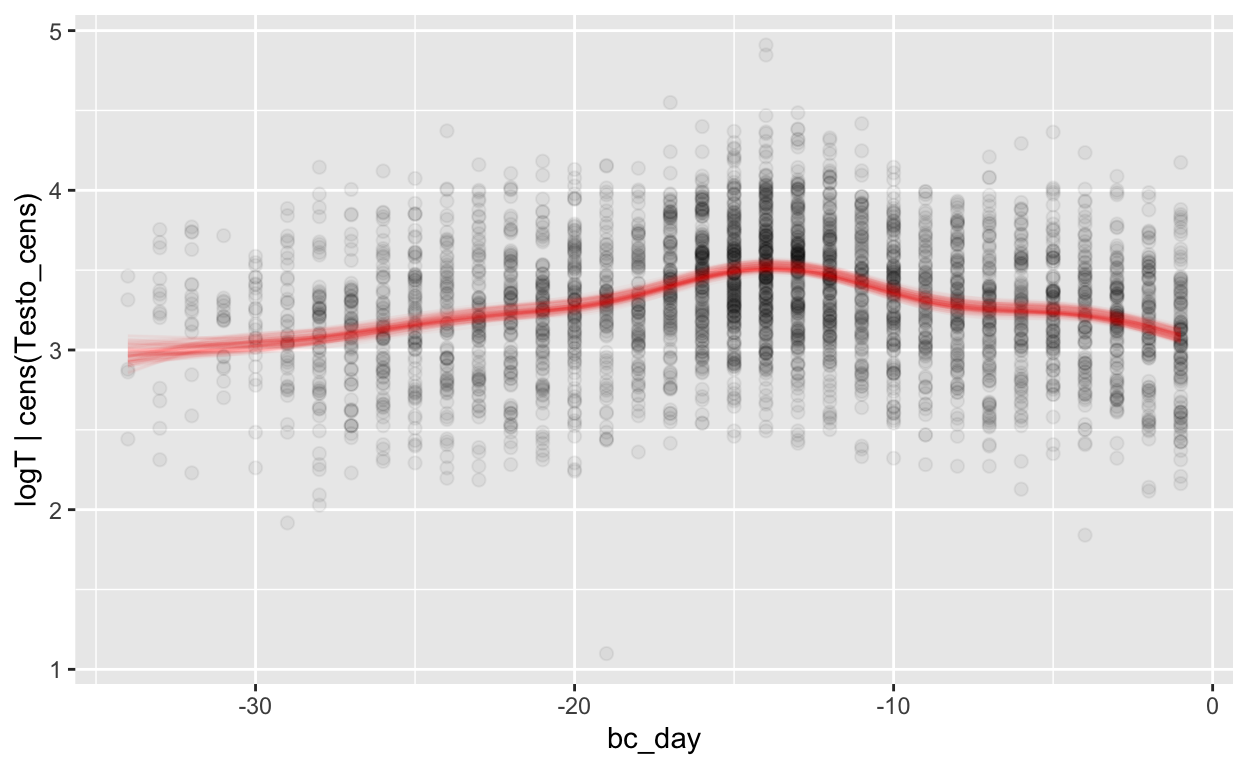
Forward counting
mod_t_fc = brm(bf(logT | cens(Testo_cens) ~ s(fc_day) + (1 | id) + (1|id:cycle)),
data = biocycle, control = list(adapt_delta = 0.95, max_treedepth = 20),
file ='models/mod_t_fc')
Warning: Rows containing NAs were excluded from the model.plot_curve(mod_t_fc, method = "fitted")
Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.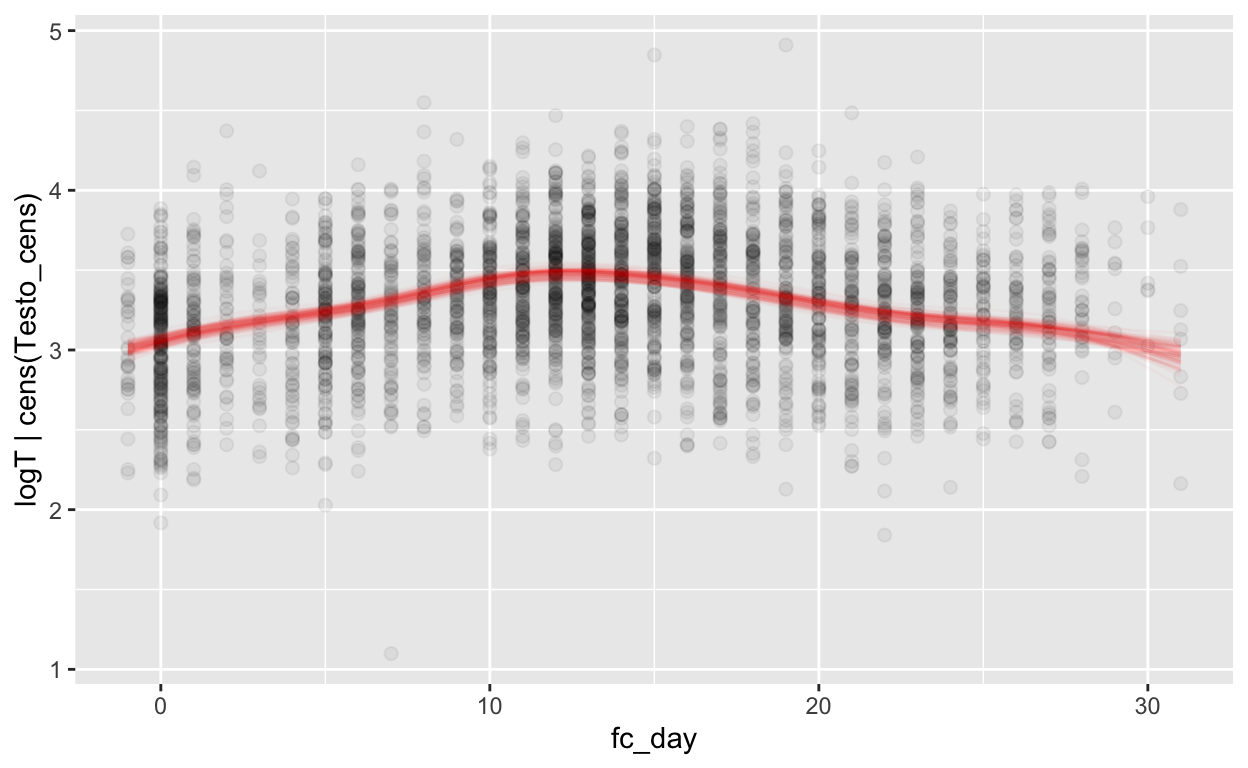
Relative to LH surge
mod_t_lh = brm(bf(logT | cens(Testo_cens) ~ s(lh_day) + (1 | id) + (1|id:cycle)),
data = biocycle %>% filter(between(lh_day, -20, 14)), control = list(adapt_delta = 0.95, max_treedepth = 20),
file ='models/mod_t_lh')
Warning: Rows containing NAs were excluded from the model.plot_curve(mod_t_lh, method = "fitted")
Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.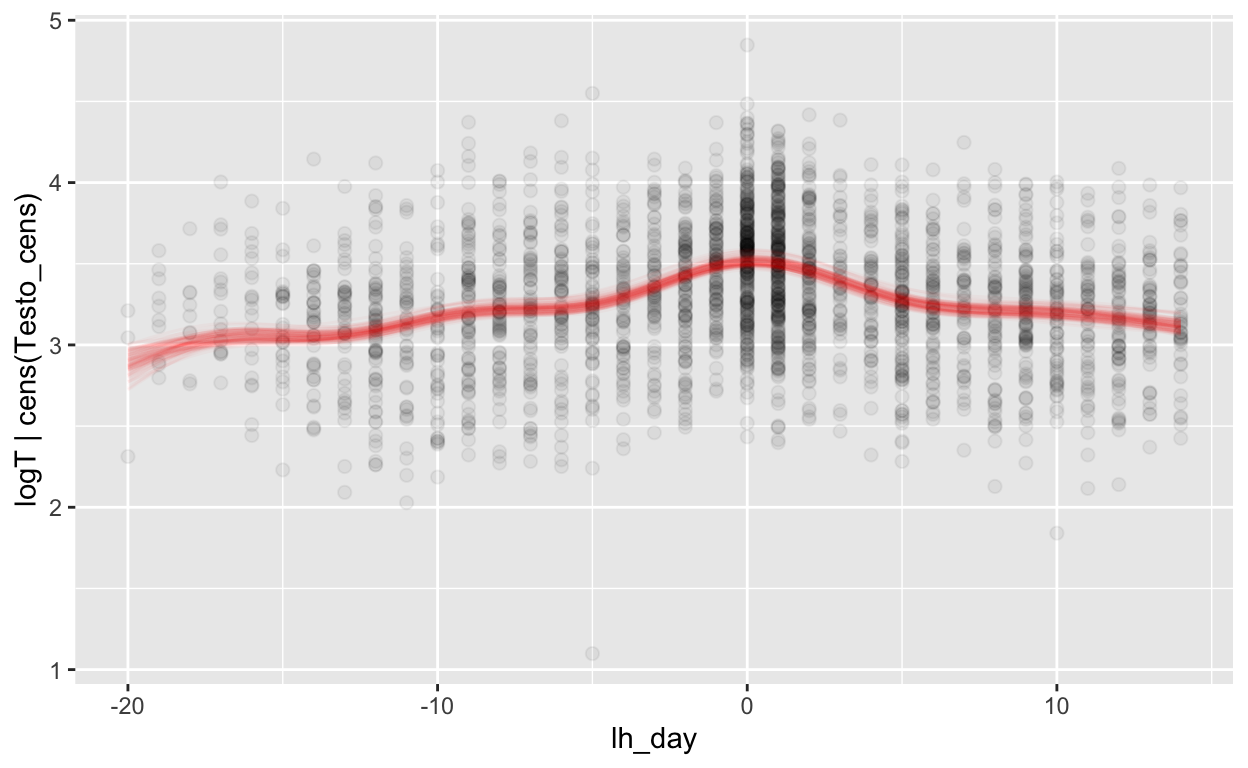
Impute FE
Backward counting
mod_ft_bc = brm(bf(logFT | cens(Testo_cens) ~ s(bc_day) + (1 | id) + (1|id:cycle)),
data = biocycle, control = list(adapt_delta = 0.95, max_treedepth = 20),
file ='models/mod_ft_bc')
Warning: Rows containing NAs were excluded from the model.plot_curve(mod_ft_bc, method = "fitted")
Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.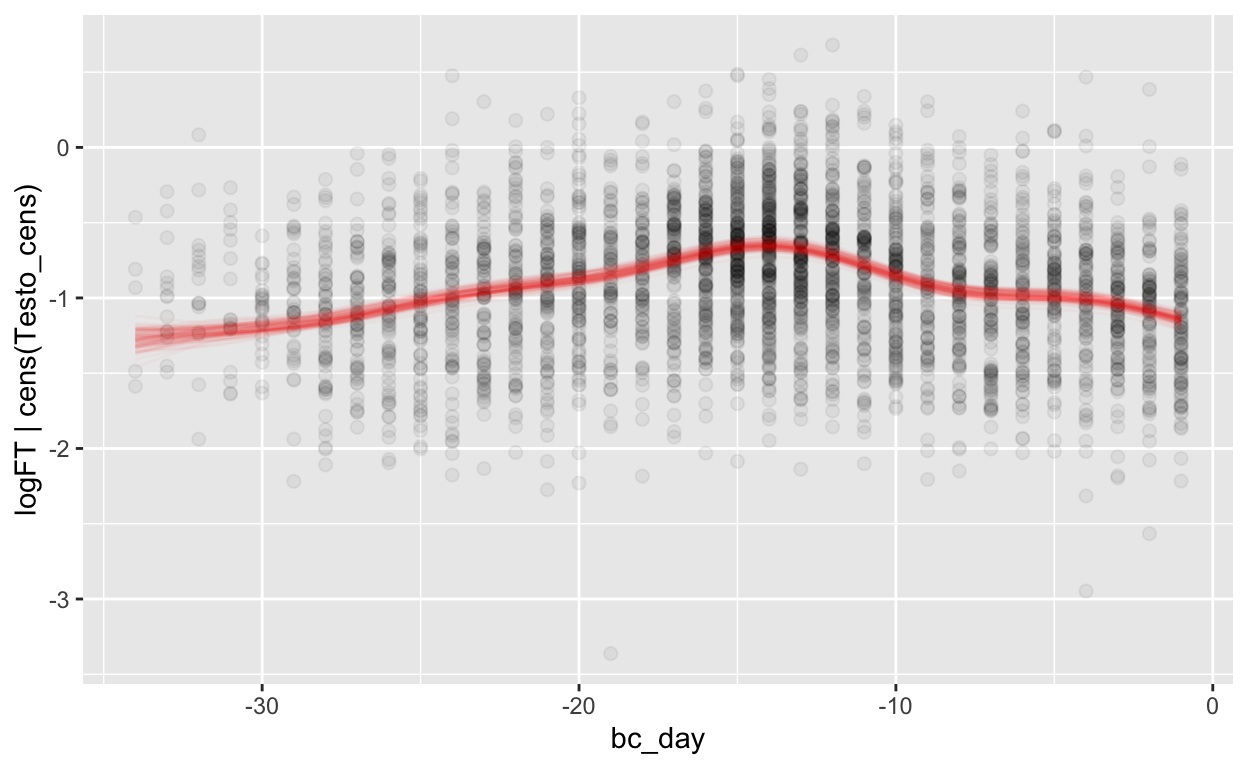
Forward counting
mod_ft_fc = brm(bf(logFT | cens(Testo_cens) ~ s(fc_day) + (1 | id) + (1|id:cycle)),
data = biocycle, control = list(adapt_delta = 0.95, max_treedepth = 20),
file ='models/mod_ft_fc')
Warning: Rows containing NAs were excluded from the model.plot_curve(mod_ft_fc, method = "fitted")
Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.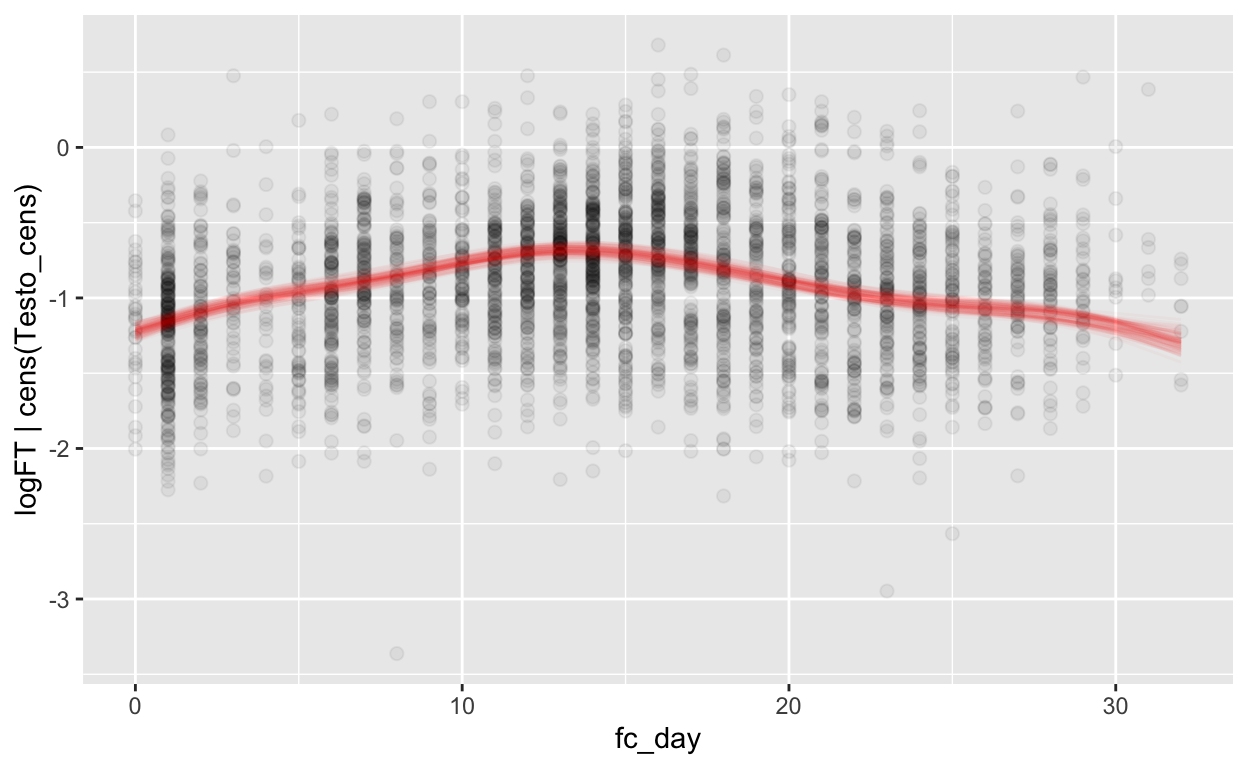
Relative to LH surge
mod_ft_lh = brm(bf(logFT | cens(Testo_cens) ~ s(lh_day) + (1 | id) + (1|id:cycle)),
data = biocycle %>% filter(between(lh_day, -20, 14)), control = list(adapt_delta = 0.95, max_treedepth = 20),
file ='models/mod_ft_lh')
Warning: Rows containing NAs were excluded from the model.plot_curve(mod_ft_lh, method = "fitted")
Warning: Argument 'nsamples' is deprecated. Please use argument
'ndraws' instead.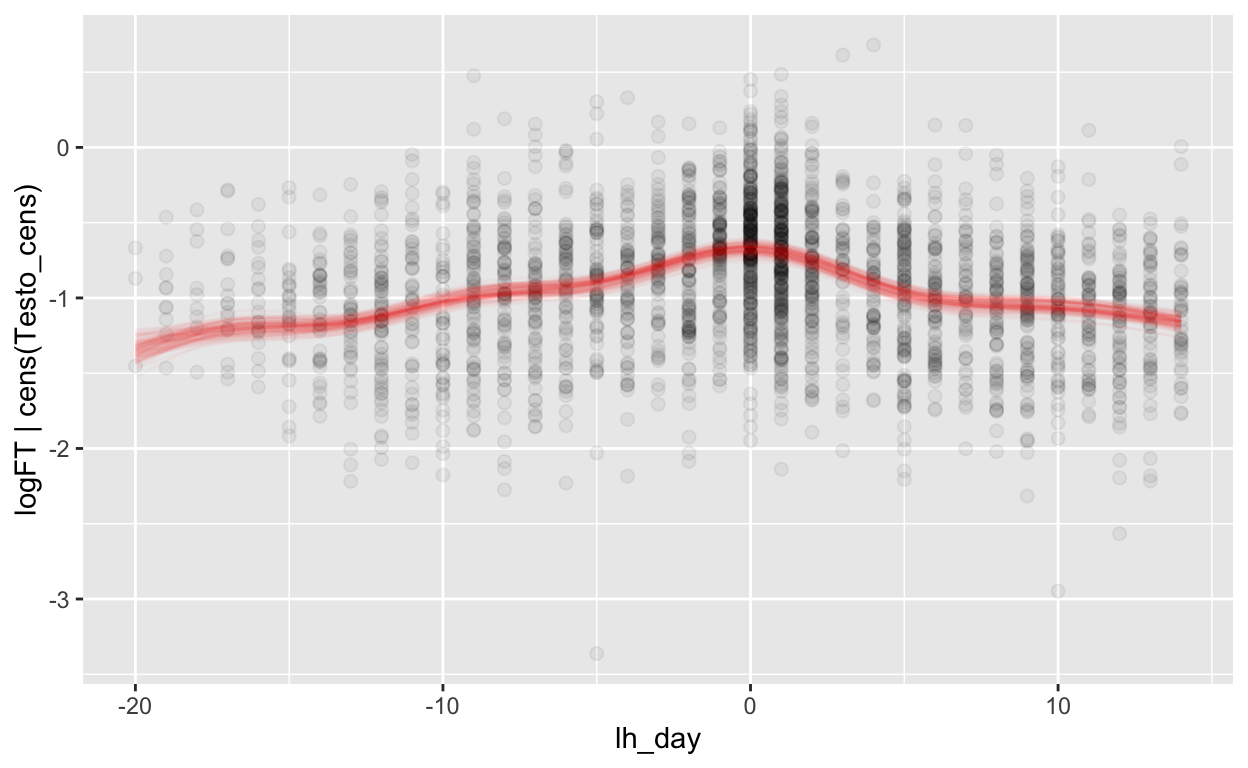
Label variables
lh_days <- as.data.frame(lh_days)
var_label(fc_days) <- list(fc_day = "Forward-counted cycle day. Counted from last (recalled) menstrual onset, starting at zero.",
prc_stirn_fc = "Probability of being in the fertile window determined according to the forward-counted cycle days, probabilities from Gangestad et al. 2016 based on Stirnemann et al. 2013",
prc_wcx_fc = "Probability of being in the fertile window determined according to the forward-counted cycle days, probabilities from Gangestad et al. 2016 based on Wilcox et al. 2001",
est_estradiol_fc = paste("Imputed serum estradiol level based on the BioCycle study, loo-R =", loo_ci(mod_e_fc)),
est_estradiol_fc_se = "Standard error: Imputed serum estradiol.",
est_estradiol_fc_low = "Lower 95% CI: Imputed serum estradiol.",
est_estradiol_fc_high = "Upper 95% CI: Imputed serum estradiol.",
est_free_estradiol_fc = paste("Imputed free serum estradiol level based on the BioCycle study, loo-R =", loo_ci(mod_fe_fc)),
est_free_estradiol_fc_se = "Standard error: Imputed free serum estradiol.",
est_free_estradiol_fc_low = "Lower 95% CI: Imputed free serum estradiol.",
est_free_estradiol_fc_high = "Upper 95% CI: Imputed free serum estradiol.",
est_progesterone_fc = paste("Imputed serum progesterone level based on the BioCycle study, loo-R =", loo_ci(mod_p_fc)),
est_progesterone_fc_se = "Standard error: Imputed serum progesterone",
est_progesterone_fc_low = "Lower 95% CI: Imputed serum progesterone",
est_progesterone_fc_high = "Upper 95% CI: Imputed serum progesterone",
est_testosterone_fc = paste("Imputed serum testosterone level based on the BioCycle study, loo-R =", loo_ci(mod_t_fc)),
est_testosterone_fc_se = "Standard error: Imputed serum testosterone",
est_testosterone_fc_low = "Lower 95% CI: Imputed serum testosterone",
est_testosterone_fc_high = "Upper 95% CI: Imputed serum testosterone",
est_free_testosterone_fc = paste("Imputed free serum testosterone level based on the BioCycle study, loo-R =", loo_ci(mod_ft_fc)),
est_free_testosterone_fc_se = "Standard error: Imputed free serum testosterone",
est_free_testosterone_fc_low = "Lower 95% CI: Imputed free serum testosterone",
est_free_testosterone_fc_high = "Upper 95% CI: Imputed free serum testosterone",
est_pbfw_fc = paste("Imputed probability of being in the fertile window based on the BioCycle study (LH test), loo-R =", loo_ci(mod_pbfw_fc)),
est_pbfw_fc_se = "Standard error: Imputed probability of being in the fertile window",
est_pbfw_fc_low = "Lower 95% CI: Imputed probability of being in the fertile window",
est_pbfw_fc_high = "Upper 95% CI: Imputed probability of being in the fertile window"
)
Warning: Results may not be meaningful for censored models.
Warning: Results may not be meaningful for censored models.
Warning: Results may not be meaningful for censored models.
Warning: Results may not be meaningful for censored models.Warning: Some Pareto k diagnostic values are too high. See help('pareto-k-diagnostic') for details.Warning: Results may not be meaningful for censored models.Warning: Some Pareto k diagnostic values are too high. See help('pareto-k-diagnostic') for details.var_label(bc_days) <- list(bc_day = "Backward-counted cycle day. Counted from next observed menstrual onset (0), beginning with -1.",
prc_stirn_bc = "Probability of being in the fertile window determined according to the backward-counted cycle days, probabilities from Gangestad et al. 2016 based on Stirnemann et al. 2013",
prc_wcx_bc = "Probability of being in the fertile window determined according to the backward-counted cycle days, probabilities from Gangestad et al. 2016 based on Wilcox et al. 2001",
est_estradiol_bc = paste("Imputed serum estradiol level based on the BioCycle study, loo-R =", loo_ci(mod_e_bc)),
est_estradiol_bc_se = "Standard error: Imputed serum estradiol.",
est_estradiol_bc_low = "Lower 95% CI: Imputed serum estradiol.",
est_estradiol_bc_high = "Upper 95% CI: Imputed serum estradiol.",
est_free_estradiol_bc = paste("Imputed free serum estradiol level based on the BioCycle study, loo-R =", loo_ci(mod_fe_bc)),
est_free_estradiol_bc_se = "Standard error: Imputed free serum estradiol.",
est_free_estradiol_bc_low = "Lower 95% CI: Imputed free serum estradiol.",
est_free_estradiol_bc_high = "Upper 95% CI: Imputed free serum estradiol.",
est_progesterone_bc = paste("Imputed serum progesterone level based on the BioCycle study, loo-R =", loo_ci(mod_p_bc)),
est_progesterone_bc_se = "Standard error: Imputed serum progesterone",
est_progesterone_bc_low = "Lower 95% CI: Imputed serum progesterone",
est_progesterone_bc_high = "Upper 95% CI: Imputed serum progesterone",
est_testosterone_bc = paste("Imputed serum testosterone level based on the BioCycle study, loo-R =", loo_ci(mod_t_bc)),
est_testosterone_bc_se = "Standard error: Imputed serum testosterone",
est_testosterone_bc_low = "Lower 95% CI: Imputed serum testosterone",
est_testosterone_bc_high = "Upper 95% CI: Imputed serum testosterone",
est_free_testosterone_bc = paste("Imputed free serum testosterone level based on the BioCycle study, loo-R =", loo_ci(mod_ft_bc)),
est_free_testosterone_bc_se = "Standard error: Imputed free serum testosterone",
est_free_testosterone_bc_low = "Lower 95% CI: Imputed free serum testosterone",
est_free_testosterone_bc_high = "Upper 95% CI: Imputed free serum testosterone",
est_pbfw_bc = paste("Imputed probability of being in the fertile window based on the BioCycle study (LH test), loo-R =", loo_ci(mod_pbfw_bc)),
est_pbfw_bc_se = "Standard error: Imputed probability of being in the fertile window",
est_pbfw_bc_low = "Lower 95% CI: Imputed probability of being in the fertile window",
est_pbfw_bc_high = "Upper 95% CI: Imputed probability of being in the fertile window"
)
Warning: Results may not be meaningful for censored models.Warning: Results may not be meaningful for censored models.
Warning: Results may not be meaningful for censored models.
Warning: Results may not be meaningful for censored models.Warning: Some Pareto k diagnostic values are too high. See help('pareto-k-diagnostic') for details.Warning: Results may not be meaningful for censored models.Warning: Some Pareto k diagnostic values are too high. See help('pareto-k-diagnostic') for details.var_label(lh_days) <- list(lh_day = "Day relative to LH surge",
est_estradiol_lh = paste("Imputed serum estradiol level based on the BioCycle study, loo-R =", loo_ci(mod_e_lh)),
est_estradiol_lh_se = "Standard error: Imputed serum estradiol.",
est_estradiol_lh_low = "Lower 95% CI: Imputed serum estradiol.",
est_estradiol_lh_high = "Upper 95% CI: Imputed serum estradiol.",
est_free_estradiol_lh = paste("Imputed free serum estradiol level based on the BioCycle study, loo-R =", loo_ci(mod_fe_lh)),
est_free_estradiol_lh_se = "Standard error: Imputed free serum estradiol.",
est_free_estradiol_lh_low = "Lower 95% CI: Imputed free serum estradiol.",
est_free_estradiol_lh_high = "Upper 95% CI: Imputed free serum estradiol.",
est_progesterone_lh = paste("Imputed serum progesterone level based on the BioCycle study, loo-R =", loo_ci(mod_p_lh)),
est_progesterone_lh_se = "Standard error: Imputed serum progesterone",
est_progesterone_lh_low = "Lower 95% CI: Imputed serum progesterone",
est_progesterone_lh_high = "Upper 95% CI: Imputed serum progesterone",
est_testosterone_lh = paste("Imputed serum testosterone level based on the BioCycle study, loo-R =", loo_ci(mod_t_lh)),
est_testosterone_lh_se = "Standard error: Imputed serum testosterone",
est_testosterone_lh_low = "Lower 95% CI: Imputed serum testosterone",
est_testosterone_lh_high = "Upper 95% CI: Imputed serum testosterone",
est_free_testosterone_lh = paste("Imputed free serum testosterone level based on the BioCycle study, loo-R =", loo_ci(mod_ft_lh)),
est_free_testosterone_lh_se = "Standard error: Imputed free serum testosterone",
est_free_testosterone_lh_low = "Lower 95% CI: Imputed free serum testosterone",
est_free_testosterone_lh_high = "Upper 95% CI: Imputed free serum testosterone",
conception_risk_lh = "Risk of conception according to Stern, Kordsmeyer, & Penke 2021 Table 1 weighted averages.",
fertile_lh = "Probability of being in the fertile window according to Stern, Kordsmeyer, & Penke 2021 Table 1, taking highest conception risk as the denominator and brought to same mean as Stirnemann numbers."
)
Warning: Results may not be meaningful for censored models.Warning: Results may not be meaningful for censored models.
Warning: Results may not be meaningful for censored models.
Warning: Results may not be meaningful for censored models.Warning: Some Pareto k diagnostic values are too high. See help('pareto-k-diagnostic') for details.Warning: Results may not be meaningful for censored models.Warning: Some Pareto k diagnostic values are too high. See help('pareto-k-diagnostic') for details.Save merge files
rio::export(fc_days, "merge_files/fc_days.rds")
rio::export(bc_days, "merge_files/bc_days.rds")
rio::export(lh_days, "merge_files/lh_days.rds")
rio::export(fc_days, "merge_files/fc_days.sav")
rio::export(bc_days, "merge_files/bc_days.sav")
rio::export(lh_days, "merge_files/lh_days.sav")
rio::export(fc_days, "merge_files/fc_days.tsv")
rio::export(bc_days, "merge_files/bc_days.tsv")
rio::export(lh_days, "merge_files/lh_days.tsv")
Codebooks
Forward-counted
codebook::compact_codebook(fc_days)
Metadata
Description
Dataset name: results
The dataset has N=40 rows and 27 columns. 40 rows have no missing values on any column.
Metadata for search engines
- Date published: 2022-03-10
|
Codebook table
JSON-LD metadata
The following JSON-LD can be found by search engines, if you share this codebook publicly on the web.
{
"name": "results",
"datePublished": "2022-03-10",
"description": "The dataset has N=40 rows and 27 columns.\n40 rows have no missing values on any column.\n\n\n## Table of variables\nThis table contains variable names, labels, and number of missing values.\nSee the complete codebook for more.\n\n[truncated]\n\n### Note\nThis dataset was automatically described using the [codebook R package](https://rubenarslan.github.io/codebook/) (version 0.9.2).",
"keywords": ["fc_day", "prc_stirn_fc", "prc_wcx_fc", "est_estradiol_fc", "est_estradiol_fc_se", "est_estradiol_fc_low", "est_estradiol_fc_high", "est_free_estradiol_fc", "est_free_estradiol_fc_se", "est_free_estradiol_fc_low", "est_free_estradiol_fc_high", "est_progesterone_fc", "est_progesterone_fc_se", "est_progesterone_fc_low", "est_progesterone_fc_high", "est_pbfw_fc", "est_pbfw_fc_se", "est_pbfw_fc_low", "est_pbfw_fc_high", "est_testosterone_fc", "est_testosterone_fc_se", "est_testosterone_fc_low", "est_testosterone_fc_high", "est_free_testosterone_fc", "est_free_testosterone_fc_se", "est_free_testosterone_fc_low", "est_free_testosterone_fc_high"],
"@context": "http://schema.org/",
"@type": "Dataset",
"variableMeasured": [
{
"name": "fc_day",
"description": "Forward-counted cycle day. Counted from last (recalled) menstrual onset, starting at zero.",
"@type": "propertyValue"
},
{
"name": "prc_stirn_fc",
"description": "Probability of being in the fertile window determined according to the forward-counted cycle days, probabilities from Gangestad et al. 2016 based on Stirnemann et al. 2013",
"@type": "propertyValue"
},
{
"name": "prc_wcx_fc",
"description": "Probability of being in the fertile window determined according to the forward-counted cycle days, probabilities from Gangestad et al. 2016 based on Wilcox et al. 2001",
"@type": "propertyValue"
},
{
"name": "est_estradiol_fc",
"description": "Imputed serum estradiol level based on the BioCycle study, loo-R = 0.57 [0.55;0.60]",
"@type": "propertyValue"
},
{
"name": "est_estradiol_fc_se",
"description": "Standard error: Imputed serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_estradiol_fc_low",
"description": "Lower 95% CI: Imputed serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_estradiol_fc_high",
"description": "Upper 95% CI: Imputed serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_free_estradiol_fc",
"description": "Imputed free serum estradiol level based on the BioCycle study, loo-R = 0.57 [0.55;0.59]",
"@type": "propertyValue"
},
{
"name": "est_free_estradiol_fc_se",
"description": "Standard error: Imputed free serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_free_estradiol_fc_low",
"description": "Lower 95% CI: Imputed free serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_free_estradiol_fc_high",
"description": "Upper 95% CI: Imputed free serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_progesterone_fc",
"description": "Imputed serum progesterone level based on the BioCycle study, loo-R = 0.74 [0.72;0.76]",
"@type": "propertyValue"
},
{
"name": "est_progesterone_fc_se",
"description": "Standard error: Imputed serum progesterone",
"@type": "propertyValue"
},
{
"name": "est_progesterone_fc_low",
"description": "Lower 95% CI: Imputed serum progesterone",
"@type": "propertyValue"
},
{
"name": "est_progesterone_fc_high",
"description": "Upper 95% CI: Imputed serum progesterone",
"@type": "propertyValue"
},
{
"name": "est_pbfw_fc",
"description": "Imputed probability of being in the fertile window based on the BioCycle study (LH test), loo-R = 0.61 [0.58;0.63]",
"@type": "propertyValue"
},
{
"name": "est_pbfw_fc_se",
"description": "Standard error: Imputed probability of being in the fertile window",
"@type": "propertyValue"
},
{
"name": "est_pbfw_fc_low",
"description": "Lower 95% CI: Imputed probability of being in the fertile window",
"@type": "propertyValue"
},
{
"name": "est_pbfw_fc_high",
"description": "Upper 95% CI: Imputed probability of being in the fertile window",
"@type": "propertyValue"
},
{
"name": "est_testosterone_fc",
"description": "Imputed serum testosterone level based on the BioCycle study, loo-R = 0.23 [0.18;0.27]",
"@type": "propertyValue"
},
{
"name": "est_testosterone_fc_se",
"description": "Standard error: Imputed serum testosterone",
"@type": "propertyValue"
},
{
"name": "est_testosterone_fc_low",
"description": "Lower 95% CI: Imputed serum testosterone",
"@type": "propertyValue"
},
{
"name": "est_testosterone_fc_high",
"description": "Upper 95% CI: Imputed serum testosterone",
"@type": "propertyValue"
},
{
"name": "est_free_testosterone_fc",
"description": "Imputed free serum testosterone level based on the BioCycle study, loo-R = 0.18 [0.10;0.23]",
"@type": "propertyValue"
},
{
"name": "est_free_testosterone_fc_se",
"description": "Standard error: Imputed free serum testosterone",
"@type": "propertyValue"
},
{
"name": "est_free_testosterone_fc_low",
"description": "Lower 95% CI: Imputed free serum testosterone",
"@type": "propertyValue"
},
{
"name": "est_free_testosterone_fc_high",
"description": "Upper 95% CI: Imputed free serum testosterone",
"@type": "propertyValue"
}
]
}`Backward-counted
codebook::compact_codebook(bc_days)
Metadata
Description
Dataset name: results
The dataset has N=40 rows and 27 columns. 40 rows have no missing values on any column.
Metadata for search engines
- Date published: 2022-03-10
|
Codebook table
JSON-LD metadata
The following JSON-LD can be found by search engines, if you share this codebook publicly on the web.
{
"name": "results",
"datePublished": "2022-03-10",
"description": "The dataset has N=40 rows and 27 columns.\n40 rows have no missing values on any column.\n\n\n## Table of variables\nThis table contains variable names, labels, and number of missing values.\nSee the complete codebook for more.\n\n[truncated]\n\n### Note\nThis dataset was automatically described using the [codebook R package](https://rubenarslan.github.io/codebook/) (version 0.9.2).",
"keywords": ["bc_day", "prc_stirn_bc", "prc_wcx_bc", "est_estradiol_bc", "est_estradiol_bc_se", "est_estradiol_bc_low", "est_estradiol_bc_high", "est_free_estradiol_bc", "est_free_estradiol_bc_se", "est_free_estradiol_bc_low", "est_free_estradiol_bc_high", "est_progesterone_bc", "est_progesterone_bc_se", "est_progesterone_bc_low", "est_progesterone_bc_high", "est_pbfw_bc", "est_pbfw_bc_se", "est_pbfw_bc_low", "est_pbfw_bc_high", "est_testosterone_bc", "est_testosterone_bc_se", "est_testosterone_bc_low", "est_testosterone_bc_high", "est_free_testosterone_bc", "est_free_testosterone_bc_se", "est_free_testosterone_bc_low", "est_free_testosterone_bc_high"],
"@context": "http://schema.org/",
"@type": "Dataset",
"variableMeasured": [
{
"name": "bc_day",
"description": "Backward-counted cycle day. Counted from next observed menstrual onset (0), beginning with -1.",
"@type": "propertyValue"
},
{
"name": "prc_stirn_bc",
"description": "Probability of being in the fertile window determined according to the backward-counted cycle days, probabilities from Gangestad et al. 2016 based on Stirnemann et al. 2013",
"@type": "propertyValue"
},
{
"name": "prc_wcx_bc",
"description": "Probability of being in the fertile window determined according to the backward-counted cycle days, probabilities from Gangestad et al. 2016 based on Wilcox et al. 2001",
"@type": "propertyValue"
},
{
"name": "est_estradiol_bc",
"description": "Imputed serum estradiol level based on the BioCycle study, loo-R = 0.68 [0.66;0.69]",
"@type": "propertyValue"
},
{
"name": "est_estradiol_bc_se",
"description": "Standard error: Imputed serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_estradiol_bc_low",
"description": "Lower 95% CI: Imputed serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_estradiol_bc_high",
"description": "Upper 95% CI: Imputed serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_free_estradiol_bc",
"description": "Imputed free serum estradiol level based on the BioCycle study, loo-R = 0.68 [0.66;0.69]",
"@type": "propertyValue"
},
{
"name": "est_free_estradiol_bc_se",
"description": "Standard error: Imputed free serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_free_estradiol_bc_low",
"description": "Lower 95% CI: Imputed free serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_free_estradiol_bc_high",
"description": "Upper 95% CI: Imputed free serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_progesterone_bc",
"description": "Imputed serum progesterone level based on the BioCycle study, loo-R = 0.83 [0.81;0.84]",
"@type": "propertyValue"
},
{
"name": "est_progesterone_bc_se",
"description": "Standard error: Imputed serum progesterone",
"@type": "propertyValue"
},
{
"name": "est_progesterone_bc_low",
"description": "Lower 95% CI: Imputed serum progesterone",
"@type": "propertyValue"
},
{
"name": "est_progesterone_bc_high",
"description": "Upper 95% CI: Imputed serum progesterone",
"@type": "propertyValue"
},
{
"name": "est_pbfw_bc",
"description": "Imputed probability of being in the fertile window based on the BioCycle study (LH test), loo-R = 0.69 [0.66;0.71]",
"@type": "propertyValue"
},
{
"name": "est_pbfw_bc_se",
"description": "Standard error: Imputed probability of being in the fertile window",
"@type": "propertyValue"
},
{
"name": "est_pbfw_bc_low",
"description": "Lower 95% CI: Imputed probability of being in the fertile window",
"@type": "propertyValue"
},
{
"name": "est_pbfw_bc_high",
"description": "Upper 95% CI: Imputed probability of being in the fertile window",
"@type": "propertyValue"
},
{
"name": "est_testosterone_bc",
"description": "Imputed serum testosterone level based on the BioCycle study, loo-R = 0.24 [0.19;0.28]",
"@type": "propertyValue"
},
{
"name": "est_testosterone_bc_se",
"description": "Standard error: Imputed serum testosterone",
"@type": "propertyValue"
},
{
"name": "est_testosterone_bc_low",
"description": "Lower 95% CI: Imputed serum testosterone",
"@type": "propertyValue"
},
{
"name": "est_testosterone_bc_high",
"description": "Upper 95% CI: Imputed serum testosterone",
"@type": "propertyValue"
},
{
"name": "est_free_testosterone_bc",
"description": "Imputed free serum testosterone level based on the BioCycle study, loo-R = 0.22 [0.17;0.27]",
"@type": "propertyValue"
},
{
"name": "est_free_testosterone_bc_se",
"description": "Standard error: Imputed free serum testosterone",
"@type": "propertyValue"
},
{
"name": "est_free_testosterone_bc_low",
"description": "Lower 95% CI: Imputed free serum testosterone",
"@type": "propertyValue"
},
{
"name": "est_free_testosterone_bc_high",
"description": "Upper 95% CI: Imputed free serum testosterone",
"@type": "propertyValue"
}
]
}`Relative to LH surge
codebook::compact_codebook(lh_days)
Metadata
Description
Dataset name: results
The dataset has N=31 rows and 23 columns. 31 rows have no missing values on any column.
Metadata for search engines
- Date published: 2022-03-10
|
Codebook table
JSON-LD metadata
The following JSON-LD can be found by search engines, if you share this codebook publicly on the web.
{
"name": "results",
"datePublished": "2022-03-10",
"description": "The dataset has N=31 rows and 23 columns.\n31 rows have no missing values on any column.\n\n\n## Table of variables\nThis table contains variable names, labels, and number of missing values.\nSee the complete codebook for more.\n\n[truncated]\n\n### Note\nThis dataset was automatically described using the [codebook R package](https://rubenarslan.github.io/codebook/) (version 0.9.2).",
"keywords": ["conception_risk_lh", "lh_day", "fertile_lh", "est_estradiol_lh", "est_estradiol_lh_se", "est_estradiol_lh_low", "est_estradiol_lh_high", "est_free_estradiol_lh", "est_free_estradiol_lh_se", "est_free_estradiol_lh_low", "est_free_estradiol_lh_high", "est_progesterone_lh", "est_progesterone_lh_se", "est_progesterone_lh_low", "est_progesterone_lh_high", "est_testosterone_lh", "est_testosterone_lh_se", "est_testosterone_lh_low", "est_testosterone_lh_high", "est_free_testosterone_lh", "est_free_testosterone_lh_se", "est_free_testosterone_lh_low", "est_free_testosterone_lh_high"],
"@context": "http://schema.org/",
"@type": "Dataset",
"variableMeasured": [
{
"name": "conception_risk_lh",
"description": "Risk of conception according to Stern, Kordsmeyer, & Penke 2021 Table 1 weighted averages.",
"@type": "propertyValue"
},
{
"name": "lh_day",
"description": "Day relative to LH surge",
"@type": "propertyValue"
},
{
"name": "fertile_lh",
"description": "Probability of being in the fertile window according to Stern, Kordsmeyer, & Penke 2021 Table 1, taking highest conception risk as the denominator and brought to same mean as Stirnemann numbers.",
"@type": "propertyValue"
},
{
"name": "est_estradiol_lh",
"description": "Imputed serum estradiol level based on the BioCycle study, loo-R = 0.72 [0.70;0.74]",
"@type": "propertyValue"
},
{
"name": "est_estradiol_lh_se",
"description": "Standard error: Imputed serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_estradiol_lh_low",
"description": "Lower 95% CI: Imputed serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_estradiol_lh_high",
"description": "Upper 95% CI: Imputed serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_free_estradiol_lh",
"description": "Imputed free serum estradiol level based on the BioCycle study, loo-R = 0.72 [0.70;0.74]",
"@type": "propertyValue"
},
{
"name": "est_free_estradiol_lh_se",
"description": "Standard error: Imputed free serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_free_estradiol_lh_low",
"description": "Lower 95% CI: Imputed free serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_free_estradiol_lh_high",
"description": "Upper 95% CI: Imputed free serum estradiol.",
"@type": "propertyValue"
},
{
"name": "est_progesterone_lh",
"description": "Imputed serum progesterone level based on the BioCycle study, loo-R = 0.87 [0.85;0.88]",
"@type": "propertyValue"
},
{
"name": "est_progesterone_lh_se",
"description": "Standard error: Imputed serum progesterone",
"@type": "propertyValue"
},
{
"name": "est_progesterone_lh_low",
"description": "Lower 95% CI: Imputed serum progesterone",
"@type": "propertyValue"
},
{
"name": "est_progesterone_lh_high",
"description": "Upper 95% CI: Imputed serum progesterone",
"@type": "propertyValue"
},
{
"name": "est_testosterone_lh",
"description": "Imputed serum testosterone level based on the BioCycle study, loo-R = 0.27 [0.22;0.31]",
"@type": "propertyValue"
},
{
"name": "est_testosterone_lh_se",
"description": "Standard error: Imputed serum testosterone",
"@type": "propertyValue"
},
{
"name": "est_testosterone_lh_low",
"description": "Lower 95% CI: Imputed serum testosterone",
"@type": "propertyValue"
},
{
"name": "est_testosterone_lh_high",
"description": "Upper 95% CI: Imputed serum testosterone",
"@type": "propertyValue"
},
{
"name": "est_free_testosterone_lh",
"description": "Imputed free serum testosterone level based on the BioCycle study, loo-R = 0.27 [0.21;0.31]",
"@type": "propertyValue"
},
{
"name": "est_free_testosterone_lh_se",
"description": "Standard error: Imputed free serum testosterone",
"@type": "propertyValue"
},
{
"name": "est_free_testosterone_lh_low",
"description": "Lower 95% CI: Imputed free serum testosterone",
"@type": "propertyValue"
},
{
"name": "est_free_testosterone_lh_high",
"description": "Upper 95% CI: Imputed free serum testosterone",
"@type": "propertyValue"
}
]
}`